SLAC joins the global fight against COVID-19 – Part 1

As part of the international response to the COVID-19 pandemic, the U.S. Department of Energy’s SLAC National Accelerator Laboratory in Menlo Parkhas re-opened some of its facilities for essential research on the atomic structure of the virus and how it interacts with potential treatments and vaccines.
SLAC scientists are also leading the development of global standards to ensure reliable testing for the coronavirus, and they are participating in DOE working groups that are considering a wide range of proposals for coronavirus research, including high-throughput drug screening and novel approaches for building ventilators.
“Working with our fellow national labs, Stanford University and other partners, we’re applying our unique expertise and facilities to make a difference in the global fight against this disease,” said SLAC Director Chi-Chang Kao.
Targeting the virus on an atomic level
At the forefront of SLAC’s coronavirus research are techniques that use beams of X-rays or electrons to decipher how life’s molecular machinery works on the atomic level. This can help answer key questions about SARS-CoV-2, the virus that causes COVID-19, such as how it recognizes and infects cells, how it replicates within them, and how it spreads through our bodies. This detailed knowledge can feed into the development of vaccines and treatments with drugs or antibodies.
While the March 16 San Francisco Bay Area’s shelter-in-place order temporarily shut down all but essential operations at the lab, the Stanford Synchrotron Radiation Lightsource (SSRL) is operating a subset of its experimental stations to provide powerful X-rays for coronavirus-related studies.
One method they are using is X-ray crystallography, which examines biomolecules such as the spike proteins on the surface of SARS-CoV-2. These proteins dock onto receptor molecules on the surface of cells in the lungs, heart or intestines and spill the virus’s genetic material into the host cells where it replicates.
Researchers crystallize these biomolecules for analysis with SSRL’s X-rays, which produces detailed images of where each atom of the protein is located. These 3D maps can then feed into the design of targeted therapeutic interventions, for instance drugs that block spike proteins from binding to host cells.
Collaborating with researchers from other institutions, the SSRL team is working to determine the 3D structures of a number of biomolecules involved in the life cycle of SARS-CoV-2.
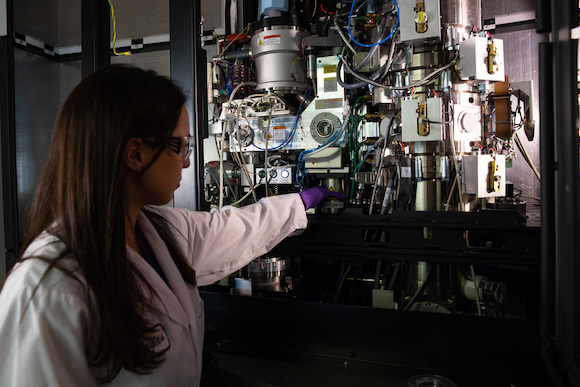
Research associate Megan Mayer loads a sample into one of SLAC’s new
“We’re able to do this research and, at the same time, follow strict social distancing protocols because our experimental stations are highly automated and require only minimal staff on site,” said Aina Cohen, who co-leads SSRL’s Structural Molecular Biology Division. “Researchers can send us frozen samples to be mounted by robots into the experimental stations. The remote researchers then use our software and video tools to control the robots, view their samples, and control all the details of their experiment from their home lab, or even from their homes.”
Another method used at SSRL is small-angle X-ray scattering, which looks at how X-rays bounce off biomolecules and their complexes in solution. Thomas Weiss, who oversees the research at this experimental station, is collaborating with Gerard Wong’s lab and other scientists at the University of California, Los Angeles, as well as researchers at the University of California, San Diego, to look at how pieces of viral proteins interact with the machinery of the human immune system. Insights into these interactions could help scientists understand why SARS-CoV-2 is so infectious and causes so much inflammation – a principal reason for why it is so deadly.
In parallel, researchers at two National Institutes of Health (NIH) centers – the Stanford-SLAC Cryo-EM Center and the National Center for Macromolecular Imaging – are pursuing answers to very similar questions with cryogenic electron microscopy (cryo-EM), which uses electron beams to study the novel coronavirus and how its molecular components interact with drugs or antibodies. One advantage of this technique is that it produces atomic-resolution images of biomolecules in their frozen, hydrated state, with no need for crystallization.
“One of the projects in full swing is a collaboration with Professor Rhiju Das at the Stanford Medical School that is aimed at designing a drug to keep viral RNA from carrying out its normal function,” said Wah Chiu, director of the NIH centers. This work involves developing synthetic RNA molecules that mimic stretches of the viral RNA, determining their 3D structures with cryo-EM and using that information to build molecules that can bind and deactivate the RNA.
In addition, Chiu and his colleagues have teamed up with several groups around the world that are preparing virus-like particles, viral proteins and RNA-protein complexes for cryo-EM analysis, as well as virus-infected cells for tomographic studies of what exactly the virus does once it enters host cells.
SLAC is also planning to bring experimental stations at its Linac Coherent Light Source (LCLS) back on line for COVID-19-related research. One of the brightest X-ray sources on Earth, this X-ray laser will complement other techniques – looking, for example, at hard-to-crystallize viral particles at room temperature to see how the virus attacks host cells and interacts with potential drugs under conditions that are close to those in the human body.
Top photo: Small-angle X-ray scattering experimental station at SSRL used for COVID-19 research. (Ivan Rajkovic/SLAC National Accelerator Laboratory); Second photo: SLAC researchers use cryogenic electron microscopy (cryo-EM) for studies of biomolecules and how they function on an atomic level. (Andy Freeberg/SLAC National Accelerator Laboratory)





